
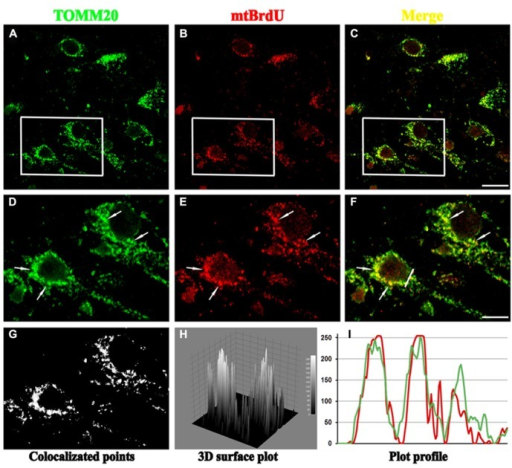
We observed 3D localizations of the individual CreS-eYFP molecules assembled into diverse filament structures extending along much of the length of the cell and across the point where the two cells divide ( Fig. P1 B). It took approximately 20 min to capture each 3D SPRAIPAINT image. We localized single Nile red molecules binding to the cell surface until a sufficient number was collected to effectively define the complex Caulobacter cell shape. Next, Nile red, a PAINT dye that binds to the membrane, was introduced to the sample. In the first stage of imaging (SPRAI), we imaged and localized single blinking eYFP molecules until the population irreversibly photobleached (i.e., was destroyed by the laser) ( Fig. P1 A). crescentus cells expressing the bacterial intermediate filament-like protein Crescentin fused to eYFP (CreS-eYFP). In our experiments, we immobilized on an agarose gel pad living C. Three-dimensional colocalization of intracellular protein structures and the cell surface with superresolution optical microscopy opens the door for the analysis of protein interactions in living cells with excellent precision (20–40 nm in 3D) over a large field of view (12 × 12 μm). Here we demonstrate live-cell 3D superresolution imaging of Crescentin-eYFP, a cytoskeletal fluorescent protein fusion, colocalized with the surface of the bacterium Caulobacter crescentus using a double-helix point spread function microscope. Simple light-induced blinking of eYFP and collisional flux onto the cell surface by Nile red are used to achieve single-molecule localizations, without any antibody labeling, cell membrane permeabilization, or thiol-oxygen scavenger systems required. We have developed a technique, termed superresolution by power-dependent active intermittency and points accumulation for imaging in nanoscale topography (SPRAIPAINT) that combines imaging of intracellular enhanced YFP (eYFP) fusions (SPRAI) with stochastic localization of the cell surface (PAINT) to image two different fluorophores sequentially with only one laser. However, live-cell superresolution imaging has been challenged by the need to image three-dimensional (3D) structures relative to their biological context, such as the cellular membrane. All rights reserved.Recently, single-molecule imaging and photocontrol have enabled superresolution optical microscopy of cellular structures beyond Abbe’s diffraction limit, extending the frontier of noninvasive imaging of structures within living cells.
Our results strongly substantiate the benefits of colocalization-based synapse detection.Įxtracellular matrix Neuron-glia co-culture Perineuronal nets Super-resolution imaging Synapse quantification.Ĭopyright © 2016 Elsevier B.V. The proposed method of synaptic protein clusters quantification exploits super-resolution imaging to provide a comprehensive approach to the analysis of postsynaptic density composition. Our Synapse Counter plugin for ImageJ offers a rapid and unbiased research tool for a broad spectrum of neuroscientists. Using super-resolution STED imaging, we demonstrate that postsynaptic puncta of completed synapses are composed of significantly more protein clusters, compared to uncompleted synapses. In a proof-of-concept study, we reveal the differential distribution of glutamatergic and GABAergic synapse density with reference to perineuronal net (PNN) expression. By combining conventional confocal microscopy with stimulated emission depletion (STED) imaging, we propose a method to quantify the number of scaffolding protein clusters, recruited to a single postsynaptic density. Structurally completed glutamatergic and GABAergic synapses were identified by VGLUT1-PSD95 and VGAT-gephyrin colocalization, respectively. Our approach is based on colocalization of pre- and postsynaptic protein markers.
Colocalization imagej software#
We present a new plugin for the open-source software ImageJ to provide a modifiable, high-throughput and easy to use method for synaptic puncta analysis. However, most of the available methods are highly tailored to specific applications and have limitations for widespread use. Thus, a number of quantitative approaches were introduced so far. Quantification of synapses and their morphological analysis are extensively used in network development and connectivity studies, drug screening and other areas of neuroscience.


 0 kommentar(er)
0 kommentar(er)
